Table of Contents
Principal Components and Linear Discriminant. Downstream Methylation Analyses with Methyl-IT
When methylation analysis is intended for diagnostic/prognostic purposes, for example, in clinical applications for patient diagnostics, to know whether the patient would be in healthy or disease stage we would like to have a good predictor tool in our side. It turns out that classical machine learning (ML) tools like hierarchical clustering, principal components and linear discriminant analysis can help us to reach such a goal. The current Methyl-IT downstream analysis is equipped with the mentioned ML tools.
1. Dataset
For the current example on methylation analysis with Methyl-IT we will use simulated data. Read-count matrices of methylated and unmethylated cytosine are generated with MethylIT.utils function simulateCounts. A basic example generating datasets is given in: Methylation analysis with Methyl-IT.
library(MethylIT) library(MethylIT.utils)
library(ggplot2)
library(ape)
alpha.ct <- 0.01
alpha.g1 <- 0.021
alpha.g2 <- 0.025
# The number of cytosine sites to generate
sites = 50000
# Set a seed for pseudo-random number generation
set.seed(124)
control.nam <- c("C1", "C2", "C3", "C4", "C5")
treatment.nam1 <- c("T1", "T2", "T3", "T4", "T5")
treatment.nam2 <- c("T6", "T7", "T8", "T9", "T10")
# Reference group
ref0 = simulateCounts(num.samples = 3, sites = sites, alpha = alpha.ct, beta = 0.5,
size = 50, theta = 4.5, sample.ids = c("R1", "R2", "R3"))
# Control group
ctrl = simulateCounts(num.samples = 5, sites = sites, alpha = alpha.ct, beta = 0.5,
size = 50, theta = 4.5, sample.ids = control.nam)
# Treatment group II
treat = simulateCounts(num.samples = 5, sites = sites, alpha = alpha.g1, beta = 0.5,
size = 50, theta = 4.5, sample.ids = treatment.nam1)
# Treatment group II
treat2 = simulateCounts(num.samples = 5, sites = sites, alpha = alpha.g2, beta = 0.5,
size = 50, theta = 4.5, sample.ids = treatment.nam2)A reference sample (virtual individual) is created using individual samples from the control population using function poolFromGRlist. The reference sample is further used to compute the information divergences of methylation levels, $TV_d$ and $H$, with function estimateDivergence [1]. This is a first fundamental step to remove the background noise (fluctuations) generated by the inherent stochasticity of the molecular processes in the cells.
# === Methylation level divergences ===
# Reference sample
ref = poolFromGRlist(ref0, stat = "mean", num.cores = 4L, verbose = FALSE)
divs <- estimateDivergence(ref = ref, indiv = c(ctrl, treat, treat2), Bayesian = TRUE,
num.cores = 6L, percentile = 1, verbose = FALSE)
# To remove hd == 0 to estimate. The methylation signal only is given for
divs = lapply(divs, function(div) div[ abs(div$hdiv) > 0 ], keep.attr = TRUE)
names(divs) <- names(divs)To get some statistical description about the sample is useful. Here, empirical critical values for the probability distribution of $H$ and $TV_d$ is obtained using quantile function from the R package stats.
critical.val <- do.call(rbind, lapply(divs, function(x) {
x <- x[x$hdiv > 0]
hd.95 = quantile(x$hdiv, 0.95)
tv.95 = quantile(abs(x$TV), 0.95)
return(c(tv = tv.95, hd = hd.95))
}))
critical.val## tv.95% hd.95%
## C1 0.2987088 21.92020
## C2 0.2916667 21.49660
## C3 0.2950820 21.71066
## C4 0.2985075 21.98416
## C5 0.3000000 22.04791
## T1 0.3376711 33.51223
## T2 0.3380282 33.00639
## T3 0.3387097 33.40514
## T4 0.3354077 31.95119
## T5 0.3402172 33.97772
## T6 0.4090909 38.05364
## T7 0.4210526 38.21258
## T8 0.4265781 38.78041
## T9 0.4084507 37.86892
## T10 0.4259411 38.607062. Modeling the methylation signal
Here, the methylation signal is expressed in terms of Hellinger divergence of methylation levels. Here, the signal distribution is modelled by a Weibull probability distribution model. Basically, the model could be a member of the generalized gamma distribution family. For example, it could be gamma distribution, Weibull, or log-normal. To describe the signal, we may prefer a model with a cross-validations: R.Cross.val > 0.95. Cross-validations for the nonlinear regressions are performed in each methylome as described in (Stevens 2009). The non-linear fit is performed through the function nonlinearFitDist.
The above statistical description of the dataset (evidently) suggests that there two main groups: control and treatments, while treatment group would split into two subgroups of samples. In the current case, to search for a good cutpoint, we do not need to use all the samples. The critical value $H_{\alpha=0.05}=33.51223$ suggests that any optimal cutpoint for the subset of samples T1 to T5 will be optimal for the samples T6 to T10 as well.
Below, we are letting the PCA+LDA model classifier to take the decision on whether a differentially methylated cytosine position is a treatment DMP. To do it, Methyl-IT function getPotentialDIMP is used to get methylation signal probabilities of the oberved $H$ values for all cytosine site (alpha = 1), in accordance with the 2-parameter Weibull distribution model. Next, this information will be used to identify DMPs using Methyl-IT function estimateCutPoint. Cytosine positions with $H$ values above the cutpoint are considered DMPs. Finally, a PCA + QDA model classifier will be fitted to classify DMPs into two classes: DMPs from control and those from treatment. Here, we fundamentally rely on a relatively strong $tv.cut \ge 0.34$ and on the signal probability distribution (nlms.wb) model.
dmps.wb <- getPotentialDIMP(LR = divs[1:10],
nlms = nlms.wb[1:10], div.col = 9L,
tv.cut = 0.34, tv.col = 7, alpha = 1,
dist.name = "Weibull2P")
cut.wb = estimateCutPoint(LR = dmps.wb, simple = FALSE,
column = c(hdiv = TRUE, TV = TRUE,
wprob = TRUE, pos = TRUE),
classifier1 = "pca.lda",
classifier2 = "pca.qda", tv.cut = 0.34,
control.names = control.nam,
treatment.names = treatment.nam1,
post.cut = 0.5, cut.values = seq(15, 38, 1),
clas.perf = TRUE, prop = 0.6,
center = FALSE, scale = FALSE,
n.pc = 4, div.col = 9L, stat = 0)
cut.wb## Cutpoint estimation with 'pca.lda' classifier
## Cutpoint search performed using model posterior probabilities
##
## Posterior probability used to get the cutpoint = 0.5
## Cytosine sites with treatment PostProbCut >= 0.5 have a
## divergence value >= 3.121796
##
## Optimized statistic: Accuracy = 1
## Cutpoint = 37.003
##
## Model classifier 'pca.qda'
##
## The accessible objects in the output list are:
## Length Class Mode
## cutpoint 1 -none- numeric
## testSetPerformance 6 confusionMatrix list
## testSetModel.FDR 1 -none- numeric
## model 2 pcaQDA list
## modelConfMatrix 6 confusionMatrix list
## initModel 1 -none- character
## postProbCut 1 -none- numeric
## postCut 1 -none- numeric
## classifier 1 -none- character
## statistic 1 -none- character
## optStatVal 1 -none- numericThe cutpoint is higher from what is expected from the higher treatment empirical critical value and DMPs are found for $H$ values: $H^{TT_{Emp}}_{\alpha=0.05}=33.98<37≤H$. The model performance in the whole dataset is:
# Model performance in in the whole dataset
cut.wb$modelConfMatrix## Confusion Matrix and Statistics
##
## Reference
## Prediction CT TT
## CT 4897 0
## TT 2 9685
##
## Accuracy : 0.9999
## 95% CI : (0.9995, 1)
## No Information Rate : 0.6641
## P-Value [Acc > NIR] : <2e-16
##
## Kappa : 0.9997
## Mcnemar's Test P-Value : 0.4795
##
## Sensitivity : 1.0000
## Specificity : 0.9996
## Pos Pred Value : 0.9998
## Neg Pred Value : 1.0000
## Prevalence : 0.6641
## Detection Rate : 0.6641
## Detection Prevalence : 0.6642
## Balanced Accuracy : 0.9998
##
## 'Positive' Class : TT
## # The False discovery rate
cut.wb$testSetModel.FDR## [1] 03. Represeting individual samples as vectors from the N-dimensional space
The above cutpoint can be used to identify DMPs from control and treatment. The PCA+QDA model classifier can be used any time to discriminate control DMPs from those treatment. DMPs are retrieved using selectDIMP function:
dmps.wb <- selectDIMP(LR = divs, div.col = 9L, cutpoint = 37, tv.cut = 0.34, tv.col = 7)Next, to represent individual samples as vectors from the N-dimensional space, we can use getGRegionsStat function from MethylIT.utils R package. Here, the simulated “chromosome” is split into regions of 450bp non-overlapping windows. and the density of Hellinger divergences values is taken for each windows.
ns <- names(dmps.wb)
DMRs <- getGRegionsStat(GR = dmps.wb, win.size = 450, step.size = 450, stat = "mean", column = 9L)
names(DMRs) <- ns4. Hierarchical Clustering
Hierarchical clustering (HC) is an unsupervised machine learning approach. HC can provide an initial estimation of the number of possible groups and information on their members. However, the effectivity of HC will depend on the experimental dataset, the metric used, and the glomeration algorithm applied. For an unknown reason (and based on the personal experience of the author working in numerical taxonomy), Ward’s agglomeration algorithm performs much better on biological experimental datasets than the rest of the available algorithms like UPGMA, UPGMC, etc.dmgm <- uniqueGRanges(DMRs, verbose = FALSE)
dmgm <- t(as.matrix(mcols(dmgm)))
rownames(dmgm) <- ns
sampleNames <- ns
hc = hclust(dist(data.frame( dmgm ))^2, method = 'ward.D')
hc.rsq = hc
hc.rsq$height <- sqrt( hc$height )4.4.1. Dendrogram
colors = sampleNames
colors[grep("C", colors)] <- "green4"
colors[grep("T[6-9]{1}", colors)] <- "red"
colors[grep("T10", colors)] <- "red"
colors[grep("T[1-5]", colors)] <- "blue"
# rgb(red, green, blue, alpha, names = NULL, maxColorValue = 1)
clusters.color = c(rgb(0, 0.7, 0, 0.1), rgb(0, 0, 1, 0.1), rgb(1, 0.2, 0, 0.1))
par(font.lab=2,font=3,font.axis=2, mar=c(0,3,2,0), family="serif" , lwd = 0.4)
plot(as.phylo(hc.rsq), tip.color = colors, label.offset = 0.5, font = 2, cex = 0.9,
edge.width = 0.4, direction = "downwards", no.margin = FALSE,
align.tip.label = TRUE, adj = 0)
axisPhylo( 2, las = 1, lwd = 0.4, cex.axis = 1.4, hadj = 0.8, tck = -0.01 )
hclust_rect(hc.rsq, k = 3L, border = c("green4", "blue", "red"),
color = clusters.color, cuts = c(0.56, 15, 0.41, 300))
Here, we have use function as.phylo from the R package ape for better dendrogram visualization and function hclust_rect from MethylIT.utils R package to draw rectangles with background colors around the branches of a dendrogram highlighting the corresponding clusters.
5. PCA + LDA
MethylIT function pcaLDA will be used to perform the PCA and PCA + LDA analyses. The function returns a list of two objects: 1) ‘lda’: an object of class ‘lda’ from package ‘MASS’. 2) ‘pca’: an object of class ‘prcomp’ from package ‘stats’. For information on how to use these objects see ?lda and ?prcomp.
Unlike hierarchical clustering (HC), LDA is a supervised machine learning approach. So, we must provide a prior classification of the individuals, which can be derived, for example, from the HC, or from a previous exploratory analysis with PCA.
# A prior classification derived from HC
grps <- cutree(hc, k = 3)
grps[grep(1, grps)] <- "CT"
grps[grep(2, grps)] <- "T1"
grps[grep(3, grps)] <- "T2"
grps <- factor(grps)
ld <- pcaLDA(data = data.frame(dmgm), grouping = grps, n.pc = 3, max.pc = 3,
scale = FALSE, center = FALSE, tol = 1e-6)
summary(ld$pca)## Importance of first k=3 (out of 15) components:
## PC1 PC2 PC3
## Standard deviation 41.5183 4.02302 3.73302
## Proportion of Variance 0.9367 0.00879 0.00757
## Cumulative Proportion 0.9367 0.94546 0.95303
We may retain enough components so that the cumulative percent of variance accounted for at least 70 to 80% [2]. By setting $scale=TRUE$ and $center=TRUE$, we could have different results and would improve or not our results. In particular, these settings are essentials if the N-dimensional space is integrated by variables from different measurement scales/units, for example, Kg and g, or Kg and Km.
5.1. PCA
The individual coordinates in the principal components (PCs) are returned by function pcaLDA. In the current case, based on the cumulative proportion of variance, the two firsts PCs carried about the 94% of the total sample variance and could split the sample into meaningful groups.
pca.coord <- ld$pca$x
pca.coord## PC1 PC2 PC3
## C1 -21.74024 0.9897934 -1.1708548 ## C2 -20.39219 -0.1583025 0.3167283 ## C3 -21.19112 0.5833411 -1.1067609 ## C4 -21.45676 -1.4534412 0.3025241 ## C5 -21.28939 0.4152275 1.0021932 ## T1 -42.81810 1.1155640 8.9577860 ## T2 -43.57967 1.1712155 2.5135643 ## T3 -42.29490 2.5326690 -0.3136530 ## T4 -40.51779 0.2819725 -1.1850555 ## T5 -44.07040 -2.6172732 -4.2384395 ## T6 -50.03354 7.5276969 -3.7333568 ## T7 -50.08428 -10.1115700 3.4624095 ## T8 -51.07915 -5.4812595 -6.7778593 ## T9 -50.27508 2.3463125 3.5371351 ## T10 -51.26195 3.5405915 -0.9489265
5.2. Graphic PC1 vs PC2
Next, the graphic for individual coordinates in the two firsts PCs can be easely visualized now:
dt <- data.frame(pca.coord[, 1:2], subgrp = grps)
p0 <- theme(
axis.text.x = element_text( face = "bold", size = 18, color="black",
# hjust = 0.5, vjust = 0.5,
family = "serif", angle = 0,
margin = margin(1,0,1,0, unit = "pt" )),
axis.text.y = element_text( face = "bold", size = 18, color="black",
family = "serif",
margin = margin( 0,0.1,0,0, unit = "mm" )),
axis.title.x = element_text(face = "bold", family = "serif", size = 18,
color="black", vjust = 0 ),
axis.title.y = element_text(face = "bold", family = "serif", size = 18,
color="black",
margin = margin( 0,2,0,0, unit = "mm" ) ),
legend.title=element_blank(),
legend.text = element_text(size = 20, face = "bold", family = "serif"),
legend.position = c(0.899, 0.12),
panel.border = element_rect(fill=NA, colour = "black",size=0.07),
panel.grid.minor = element_line(color= "white",size = 0.2),
axis.ticks = element_line(size = 0.1), axis.ticks.length = unit(0.5, "mm"),
plot.margin = unit(c(1,1,0,0), "lines"))
ggplot(dt, aes(x = PC1, y = PC2, colour = grps)) +
geom_vline(xintercept = 0, color = "white", size = 1) +
geom_hline(yintercept = 0, color = "white", size = 1) +
geom_point(size = 6) +
scale_color_manual(values = c("green4","blue","brown1")) +
stat_ellipse(aes(x = PC1, y = PC2, fill = subgrp), data = dt, type = "norm",
geom = "polygon", level = 0.5, alpha=0.2, show.legend = FALSE) +
scale_fill_manual(values = c("green4","blue","brown1")) + p0
5.3. Graphic LD1 vs LD2
In the current case, better resolution is obtained with the linear discriminant functions, which is based on the three firsts PCs. Notice that the number principal components used the LDA step must be lower than the number of individuals ($N$) divided by 3: $N/3$.
ind.coord <- predict(ld, newdata = data.frame(dmgm), type = "scores")
dt <- data.frame(ind.coord, subgrp = grps)
p0 <- theme(
axis.text.x = element_text( face = "bold", size = 18, color="black",
# hjust = 0.5, vjust = 0.5,
family = "serif", angle = 0,
margin = margin(1,0,1,0, unit = "pt" )),
axis.text.y = element_text( face = "bold", size = 18, color="black",
family = "serif",
margin = margin( 0,0.1,0,0, unit = "mm" )),
axis.title.x = element_text(face = "bold", family = "serif", size = 18,
color="black", vjust = 0 ),
axis.title.y = element_text(face = "bold", family = "serif", size = 18,
color="black",
margin = margin( 0,2,0,0, unit = "mm" ) ),
legend.title=element_blank(),
legend.text = element_text(size = 20, face = "bold", family = "serif"),
legend.position = c(0.08, 0.12),
panel.border = element_rect(fill=NA, colour = "black",size=0.07),
panel.grid.minor = element_line(color= "white",size = 0.2),
axis.ticks = element_line(size = 0.1), axis.ticks.length = unit(0.5, "mm"),
plot.margin = unit(c(1,1,0,0), "lines"))
ggplot(dt, aes(x = LD1, y = LD2, colour = grps)) +
geom_vline(xintercept = 0, color = "white", size = 1) +
geom_hline(yintercept = 0, color = "white", size = 1) +
geom_point(size = 6) +
scale_color_manual(values = c("green4","blue","brown1")) +
stat_ellipse(aes(x = LD1, y = LD2, fill = subgrp), data = dt, type = "norm",
geom = "polygon", level = 0.5, alpha=0.2, show.legend = FALSE) +
scale_fill_manual(values = c("green4","blue","brown1")) + p0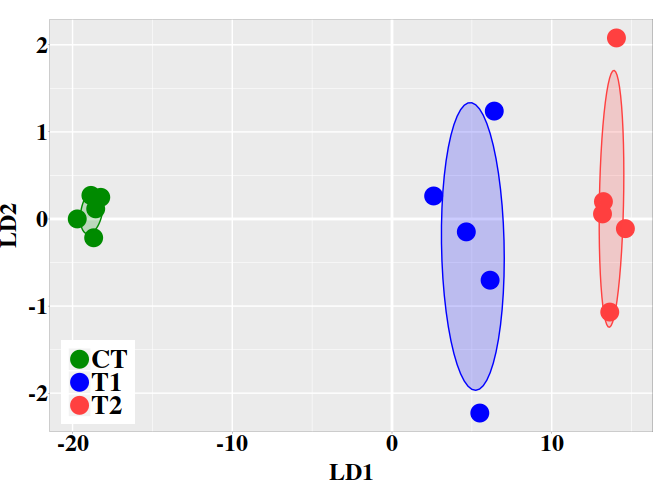
References
- Liese, Friedrich, and Igor Vajda. 2006. “On divergences and informations in statistics and information theory.” IEEE Transactions on Information Theory 52 (10): 4394–4412. doi:10.1109/TIT.2006.881731.
- Stevens, James P. 2009. Applied Multivariate Statistics for the Social Sciences. Fifth Edit. Routledge Academic.
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
Informative articles, excellent work site admin! If you’d like more information about Data Mining, drop by my site at Webemail24 Cheers to creating useful content on the web!
Great!!! Thank you for sharing this details. If you need some information about Website Traffic than have a look here Seoranko
This was a very good post. Check out my web page QN5 for additional views concerning about Thai-Massage.
Полностью актуальные события мира fashion.
Актуальные события всемирных подуимов.
Модные дома, бренды, высокая мода.
Самое приятное место для модных людей.
https://rftimes.ru/news/2024-05-22-es-utverdil-reshenie-o-konfiskatsii-pribyli-ot-aktivov-rossii-chto-eto-oznachaet
https://sevastopol.rftimes.ru/news/2024-06-13-v-sevastopole-otrazili-raketnuyu-ataku-vsu-vse-tseli-unichtozhili-v-vozduhe-postradavshih-i-razrusheniy-net
https://kursktoday.ru/news/2024-03-03-moshennik-iz-nizhnego-novgoroda-obyazan-vernut-150-tys-rubley-86-letney-pensionerke-v-kurske
https://rftimes.ru/news/2023-11-24-finskaya-politsiya-obnaruzhila-yakor-okolo-povrezhdennogo-gazoprovoda-balticconnector
https://msk.rftimes.ru/news/2024-04-17-ukrainskiy-agent-podorval-mashinu-v-moskve
startup talky Great information shared.. really enjoyed reading this post thank you author for sharing this post .. appreciated
Наиболее актуальные новинки подиума.
Исчерпывающие эвенты самых влиятельных подуимов.
Модные дома, бренды, высокая мода.
Самое приятное место для стильныех людей.
https://krasnodar.rftimes.ru/news/2024-03-03-zastrelivshego-opponenta-v-krasnodare-muzhchinu-otpravili-pod-strazhu
https://rftimes.ru/news/2024-03-05-sekrety-proizvodstva-stekol-dlya-neboskrebov-i-aeroportov
https://perm.rftimes.ru/news/2024-04-13-vosstanovleno-dvizhenie-tramvaev-v-permi-posle-obryva-kontaktnoy-seti
https://rftimes.ru/news/2023-11-24-finskaya-politsiya-obnaruzhila-yakor-okolo-povrezhdennogo-gazoprovoda-balticconnector
https://kostroma.rftimes.ru/news/2024-05-18-noch-muzeev-perenesli-iz-tsentra-kostromy-na-vystavku-svo
La weekly I do not even understand how I ended up here, but I assumed this publish used to be great
Полностью актуальные новости модного мира.
Важные новости самых влиятельных подуимов.
Модные дома, бренды, гедонизм.
Интересное место для модных людей.
https://sevastopol.rftimes.ru/news/2024-07-07-ostavshiesya-postradavshie-pri-atake-vsu-v-bolnitsah-sevastopolya-i-moskvy
https://sochidaily.ru/read/2023-11-26-zaderzhki-v-dvizhenii-passazhirskih-poezdov-v-sochi-iz-za-padeniya-derevev.html
https://kirov.rftimes.ru/news/2024-04-09-gk-interstroy-nachinaet-prodazhi-kvartir-v-zhk-ay-petri-v-yalte
https://mskfirst.ru/msk/2023-11-25-snizhenie-stoimosti-proezda-moskovskaya-kanatnaya-doroga
758https://vladnews.ru/2023-11-16/227949/demna_gvasaliya https://sochidaily.ru/read/2023-11-27-prognoziruetsya-snegopad-i-volnenie-na-more-v-sochi.html
Несомненно трендовые новости модного мира.
Важные эвенты лучших подуимов.
Модные дома, торговые марки, высокая мода.
Лучшее место для стильныех людей.
4439 https://vladnews.ru/2023-11-16/227949/demna_gvasaliya https://ekbtoday.ru/news/2024-08-05-razvitie-transportnoy-infrastruktury-ekaterinburga/
https://izhevsk.rftimes.ru/news/2024-04-23-poezd-minoborony-rossii-posetil-izhevsk
https://rftimes.ru/news/2023-11-24-boets-s-pozyvnym-shket-drony-stanut-effektivnym-sredstvom-borby-s-abrams
https://ekb.rftimes.ru/news/2024-03-01-vsplesk-nasiliya-sredi-shkolnikov-v-ekaterinburge
https://rftimes.ru/news/2024-04-07-lgotnaya-ipoteka-v-2024-vse-programmy-s-gospodderzhkoy
Полностью стильные события мира fashion.
Исчерпывающие эвенты лучших подуимов.
Модные дома, лейблы, высокая мода.
Лучшее место для трендовых людей.
https://samara.rftimes.ru/news/2024-05-30-samolet-iz-samary-posle-problem-s-shassi-prizemlilsya-v-moskve
https://omsk.rftimes.ru/news/2024-06-15-smena-pogody-poholodanie-pridet-18-iyunya
https://enovosibirsk.ru/news/2023-11-24-zaderzhan-zamestitel-direktora-departamenta-gosudarstvennoy-politiki-i-regulirovaniya-v-sfere-razvitiya-osobo-ohranyaemyh-prirodnyh-territoriy/
https://simferopol.rftimes.ru/news/2024-05-03-fotovystavka-bitva-za-krym-put-k-osvobozhdeniyu-otkrylas-v-simferopole
https://mskfirst.ru/msk/2024-01-05-vozobnovleno-teplosnabzhenie-domov-na-severo-vostoke-moskvy
BaddieHub I really like reading through a post that can make men and women think. Also, thank you for allowing me to comment!
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
Стильные советы по выбору модных видов на каждый день.
Заметки стилистов, новости, все коллекции и шоу.
https://rftimes.ru/news/2024-08-14-7-samyh-kultovyh-veshchey-ot-balenciaga
Эта вечерняя подборка событий позволит вам всегда быть информированными на тему свежих событий.
https://pitersk.ru/articles/2024-08-20-7-ocharovatelnyh-lukov-s-tsvetochnymi-platyami-zimmermann/
Наша дневная лента новостей позволит вам всегда быть информированными на тему актуальных новостей.
https://pitersk.ru/articles/2024-08-20-7-ocharovatelnyh-lukov-s-tsvetochnymi-platyami-zimmermann/
Модные заметки по подбору модных видов на каждый день.
Статьи стилистов, события, все коллекции и мероприятия.
https://vladtoday.ru/news/2024-09-10-10-prichin-za-chto-my-lyubim-demnu-gvasaliyu/
Your posts stand out from other sites I’ve read stuff from. Keep doing what you’re doing! Here, take a look at mine QH7 for content about about Airport Transfer.
Стильные заметки по выбору превосходных образов на любой день.
Обзоры профессионалов, новости, все новинки и шоу.
https://mvmedia.ru/novosti/122-gde-luchshe-pokupat-originalnye-brendovye-veshchi-kak-vybrat-nadezhnye-magaziny-i-platformy/
idlixmusicallydownpgbetpurislotbro138visa4dpulau88protogelsurgaplayhoktotopandora88operatotompoidbromo77pulsa777rajawd777onic4dzeus138jpslottumi123slot88kuwolestogelsensa138virus4didcash88mpo500topcer88casaprizetribun855sogoslotkarirtotogorila4dstake88instaslot88pvjbetsavefrom tiktokdetik288233 leyuanmpo2121giga5000snack video downloaderliveomekpantaislotelanggamebabyslotsupermpoliga788ombak1238278 slot
These are genuinely great ideas in concerning blogging. You have touched
some pleasant points here. Any way keep up wrinting.
Your article helped me a lot, is there any more related content? Thanks!
sokuja sokuja sokuja
sokuja (https://northernfortplayhouse.com/)
Its like you read my mind! You seem to know so much about this, like
you wrote the e-book in it or something. I believe that you
can do with a few p.c. to pressure the message home a bit, but instead of that,
that is excellent blog. An excellent read. I’ll
definitely be back.
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article.
Модные заметки по подбору модных луков на каждый день.
Мнения экспертов, события, все показы и мероприятия.
https://omskdaily.ru/novosti/2024-09-20-7-interesnyh-faktov-o-vetements-ot-antiglamura-do-modnogo-fenomena/
dadu online dadu online dadu online dadu online
Nice blog here! Also your web site loads
up very fast! What web host are you using? Can I get your affiliate link to
your host? I wish my web site loaded up as quickly as yours lol
uPVC Pipes in Iraq Elite Pipe Factory in Iraq provides a range of high-quality uPVC pipes, known for their durability, resistance to corrosion, and ease of installation. Our uPVC pipes are designed to meet rigorous quality standards, making them an excellent choice for a variety of applications. Recognized as one of the best and most reliable pipe manufacturers in Iraq, Elite Pipe Factory ensures that our uPVC pipes deliver outstanding performance and reliability. Learn more about our uPVC pipes by visiting elitepipeiraq.com.
rajabandot rajabandot rajabandot rajabandot rajabandot
I’m not sure where you are getting your info, but great topic.
I needs to spend some time learning more or understanding more.
Thanks for wonderful info I was looking for this info for my mission.
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
mawarslot mawarslot mawarslot mawarslot mawarslot
Does your website have a contact page? I’m having trouble locating it
but, I’d like to shoot you an e-mail. I’ve got some ideas for your blog you might
be interested in hearing. Either way, great blog and
I look forward to seeing it grow over time.
Your posts in this blog really shine! Glad to gain some new insights, which I happen to also cover on my page. Feel free to visit my webpage QU5 about Cosmetics and any tip from you will be much apreciated.
Точно стильные новости модного мира.
Все эвенты лучших подуимов.
Модные дома, лейблы, гедонизм.
Приятное место для стильныех людей.
https://outstreet.ru/yeah/11164-5-stilnyh-modeley-chasov-guess-dlya-devushki-v-2024-godu/
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
Family Dollar I like the efforts you have put in this, regards for all the great content.
hoki777 hoki777 hoki777
You actually make it seem so easy with your presentation but I
find this matter to be actually something that I think I would never understand.
It seems too complicated and extremely broad for me. I am looking forward for your next post, I will try to get the hang of it!
Hi there, I simply couldn’t leave your website without saying that I appreciate the information you supply to your visitors. Here’s mine 63U and I cover the same topic you might want to get some insights about Thai-Massage.
rüyada derenin taşması
Модные заметки по подбору стильных видов на каждый день.
Мнения профессионалов, новости, все новые коллекции и шоу.
https://luxe-moda.ru/chic/564-10-prichin-lyubit-brend-brunello-cucinelli/
Стильные советы по созданию превосходных луков на каждый день.
Заметки экспертов, новости, все коллекции и мероприятия.
https://sofiamoda.ru/style/2024-10-03-principe-di-bologna-roskosh-italyanskogo-stilya-i-elegantnost-na-kazhdyy-den/
rtp slot rtp slot rtp slot
I know this website offers quality based posts and extra material, is there any other website which gives these kinds of stuff
in quality?
I don’t think the title of your article matches the content lol. Just kidding, mainly because I had some doubts after reading the article. https://www.binance.com/ar/register?ref=V2H9AFPY
join680 join680 join680
Fantastic items from you, man. I have be mindful your stuff
previous to and you’re just too magnificent. I actually like what you have got right here, really like what you are
saying and the best way in which you say it.
You’re making it entertaining and you continue to take care of to stay it sensible.
I can’t wait to read much more from you. That is really a great site.
Бренд Balenciaga является одним из самых известных домов высокой моды, который был основан в начале 20 века известным модельером Кристобалем Баленсиагой. Он славится своими смелыми дизайнерскими решениями и неординарными формами, которые часто бросают вызов стандартам индустрии.
https://balenciaga.whitesneaker.ru/
luna togel luna togel luna togel
This site was… how do I say it? Relevant!! Finally I’ve found something that helped me.
Many thanks!
Hi there, You’ve done an excellent job. I will certainly digg it and personally recommend to my friends.
I am sure they will be benefited from this web site.
Thank you for your sharing. I am worried that I lack creative ideas. It is your article that makes me full of hope. Thank you. But, I have a question, can you help me?
Keep on writing, great job!
Hi there it’s me, I am also visiting this site
regularly, this web page is in fact fastidious and the people are genuinely sharing pleasant thoughts.
Appreciation to my father who stated to me on the topic of this website, this website is in fact awesome.
Hello there! I know this is kind of off topic but I was wondering which blog platform are you using for this site?
I’m getting fed up of WordPress because I’ve had issues with hackers and I’m looking at options for another platform.
I would be fantastic if you could point me in the direction of a good platform.
Do you have a spam problem on this website;
I also am a blogger, and I was wanting to know
your situation; we have created some nice methods and we are looking
to swap strategies with others, be sure to shoot me an email if interested.