Epigenomic research
In the course of our epigenetic research, we have been improving our statistical approaches for methylome analysis. We started with the application of the standard statistical approaches for genome-wide methylation analysis and further analysis of the previous results with the application of multivariate statistics. Our group has proposed a signal detection based approach to unveil the genome-wide methylation patterning based on the statistical physics effect of methylation on DNA molecules. Since the biological signal created within the dynamic methylome environment characteristic of plants is not free of background noise (as in all known natural or technologically generated signals), an additional application of signal detection theory is accomplished. The signal-detection based approach is postulated to provide greater sensitivity for resolving true signal from thermodynamic background “noise” within the methylome system.
Currently, we have implemented our novel methylome analysis procedure in an R package named Methyl-IT, which is available at GitHub. This approach permitted the identification of differentially methylated genes (DMGs) associated with stress response and circadian rhythm in msh1 descendant plants with segregation of the RNAi transgene. Results suggest that a mild methylation patterning persist in response to the artificial stress condition originally induced by RNAi suppression of the MSH1 gene in the ancestor plants, producing recurrently heritable memory. Downstream analysis of DMGs and differentially expressed genes (DEGs) with network based enrichment analysis identified overlapping (DMGs and DEGs) gene networks in response to the artificial stress condition.
Examples on methylation analysis with Methyl-IT, written to guide the users, are:
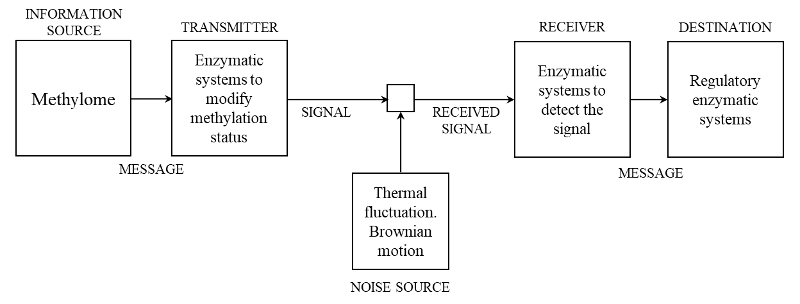
The methylation communication system. Schematic diagram of a general communication system as given by Shannon (1948) [1] and its particularization for methylation communication system (MCS). By a communication system we will mean a system of the type indicated schematically in the diagram. According with Shannon (1948), it consists of essentially five parts:
- An information source which produces a message or sequence of messages to be communicated to the receiving terminal. In the case of the MCS, it is the methylome, which is integrated by the set of all genome-wide binary stretches of methylated and non-methylated cytosines.
- A transmitter which operates on the message in some way to produce a signal suitable for transmission over the channel. In the case of MCS, it is integrated by the enzymatic systems necessary to modify the methylation status according to the requirements determined by the changes in the environmental condition. These are all the multicomponent molecular machines that methylate or demethylate the cytosine throughout different biochemical pathways.
- The channel is merely the medium used to transmit the signal from transmitter to receiver, which in the case of MCS is the DNA molecule.
- The receiver ordinarily performs the inverse operation of that done by the transmitter, reconstructing the message from the signal. In the case of MCS, it is formed by single or multicomponent molecular machines able to detect the signal on the DNA molecule. In the MCS, the receiver perform an additional task, the decoding of the signal into a message meaningful for the regulatory enzymatic machinery (also found in human communication systems). This implies the existence of a methylation code, which is the set of methylation rules that determine whether or not a binary stretch of methylation marks is a meaningful signal for recognition (a word) by the molecular machines that trigger tissue response. In addition, the enzymatic systems in charge of genome-wide modification of methylation status can be simultaneously “receiver” and “destination”, which is a function ordinarily not found in the current human technical communication systems.
- The destination is the person (or thing) for whom the message is intended. In the case of MCS, it could be the regulatory enzymatic systems or the enzymatic systems in charge of the genome-wide modification of methylation status.
Statistical physic evidence that support the existence of a MCS is given in reference [2]. Deciphering the Epigenetic Code from the Human MCS will be the biggest challenge for the current post-genomics era, the biggest breakthrough in the history of biomedical research. It will set the basis to confront/control genetic and infections diseases, aging process, etc, in very innovative ways, engineering the physiological response of living organisms to the environmental variations.
References
- Shannon C. E (1948) A Mathematical Theory of Communication. Bell Syst Tech J 27: 379–423.
- Sanchez R, Mackenzie SA. Information Thermodynamics of Cytosine DNA Methylation. PLoS One, 2016, 11:e0150427
